5UG9
Target information
- RCSB PDB
- 5UG9
- Title
- Crystal structure of the EGFR kinase domain (L858R, T790M, V948R) in complex with a covalent inhibitor N-[(3R,4R)-4-fluoro-1-{6-[(3-methoxy-1-methyl-1H-pyrazol-4-yl)amino]-9-(propan-2-yl)-9H-purin-2-yl}pyrrolidin-3-yl]propanamide
- Method
- X-RAY DIFFRACTION
- Resolution
- 1.33
- Classification
- TRANSFERASE/TRANSFERASE INHIBITOR
- Organism
- Homo sapiens
- Protein
- Epidermal growth factor receptor (P00533) Looking for covalent inhibitors of this target ?
- Year
- 2017
- Publication Title
- Discovery of N-((3R,4R)-4-Fluoro-1-(6-((3-methoxy-1-methyl-1H-pyrazol-4-yl)amino)-9-methyl-9H-purin-2-yl)pyrrolidine-3-yl)acrylamide (PF-06747775) through Structure-Based Drug Design: A High Affinity Irreversible Inhibitor Targeting Oncogenic EGFR Mutants with Selectivity over Wild-Type EGFR.
- Abstract
-
Mutant epidermal growth factor receptor (EGFR) is a major driver of non-small-cell lung cancer (NSCLC). Marketed first generation inhibitors, such as erlotinib, effect a transient beneficial response in EGFR mutant NSCLC patients before resistance mechanisms render these inhibitors ineffective. Secondary oncogenic EGFR mutations account for approximately 50% of relapses, the most common being the gatekeeper T790M substitution that renders existing therapies ineffective. The discovery of PF-06459988 (1), an irreversible pyrrolopyrimidine inhibitor of EGFR T790M mutants, was recently disclosed.1 Herein, we describe our continued efforts to achieve potency across EGFR oncogenic mutations and improved kinome selectivity, resulting in the discovery of clinical candidate PF-06747775 (21), which provides potent EGFR activity against the four common mutants (exon 19 deletion (Del), L858R, and double mutants T790M/L858R and T790M/Del), selectivity over wild-type EGFR, and desirable ADME properties. Compound 21 is currently being evaluated in phase-I clinical trials of mutant EGFR driven NSCLC.
- External Link
- RCSB PDB
Ligand information
- HET
- 8AM
- Chain ID
- A
- HET Number
- 9001
- Molecular Formula
- C20H26FN9O2
- Structure
-
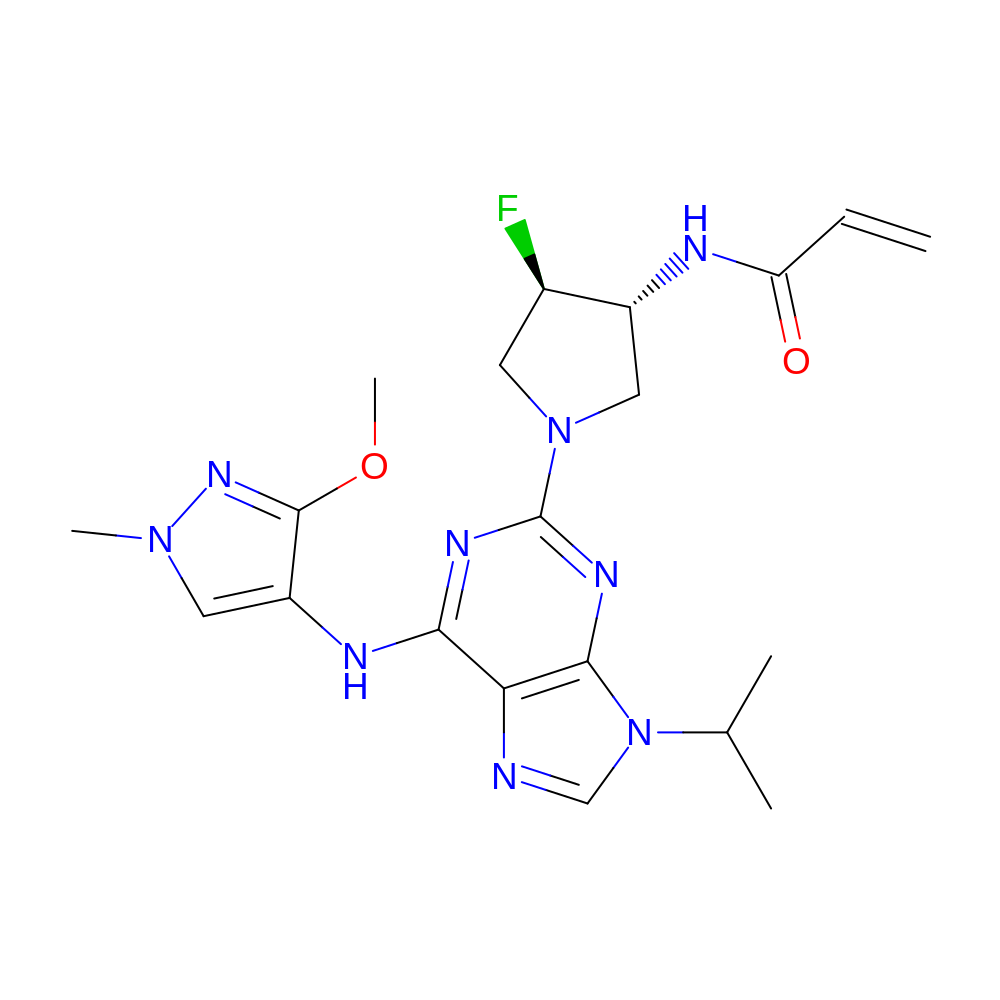
- IUPAC Name
- N-[(3R,4R)-4-fluoro-1-[9-isopropyl-6-[(3-methoxy-1-methyl-pyrazol-4-yl)amino]purin-2-yl]pyrrolidin-3-yl]prop-2-enamide
- InChI
- InChI=1S/C20H26FN9O2/c1-6-15(31)23-13-9-29(7-12(13)21)20-25-17(24-14-8-28(4)27-19(14)32-5)16-18(26-20)30(10-22-16)11(2)3/h6,8,10-13H,1,7,9H2,2-5H3,(H,23,31)(H,24,25,26)/t12-,13-/m1/s1
- InChI Key
- LJKPCPFZGRKCSR-CHWSQXEVSA-N
- Canonical SMILES
- COc1nn(C)cc1Nc2nc(nc(c23)n(cn3)C(C)C)N4C[C@@H](F)[C@@H](C4)NC(=O)C=C
- Bioactivity data
- CI004301
Covalent Binding
- Warhead
- Michael Acceptor
- Reaction Mechanism
- Michael Addition
- Residue
- CYS : 797
- Residue Chain
- A
- Interactions
- Pharmacophore Model