5P9K
Target information
- RCSB PDB
- 5P9K
- Title
- CRYSTAL STRUCTURE OF BTK with CNX 774
- Method
- X-RAY DIFFRACTION
- Resolution
- 1.28
- Classification
- TRANSFERASE
- Organism
- Homo sapiens
- Protein
- Tyrosine-protein kinase BTK (Q06187) Looking for covalent inhibitors of this target ?
- Year
- 2016
- Publication Title
- Ability of Bruton's Tyrosine Kinase Inhibitors to Sequester Y551 and Prevent Phosphorylation Determines Potency for Inhibition of Fc Receptor but not B-Cell Receptor Signaling.
- Abstract
-
Bruton's tyrosine kinase (Btk) is expressed in a variety of hematopoietic cells. Btk has been demonstrated to regulate signaling downstream of the B-cell receptor (BCR), Fc receptors (FcRs), and toll-like receptors. It has become an attractive drug target because its inhibition may provide significant efficacy by simultaneously blocking multiple disease mechanisms. Consequently, a large number of Btk inhibitors have been developed. These compounds have diverse binding modes, and both reversible and irreversible inhibitors have been developed. Reported herein, we have tested nine Btk inhibitors and characterized on a molecular level how their interactions with Btk define their ability to block different signaling pathways. By solving the crystal structures of Btk inhibitors bound to the enzyme, we discovered that the compounds can be classified by their ability to trigger sequestration of Btk residue Y551. In cells, we found that sequestration of Y551 renders it inaccessible for phosphorylation. The ability to sequester Y551 was an important determinant of potency against Fc??R signaling as Y551 sequestering compounds were more potent for inhibiting basophils and mast cells. This result was true for the inhibition of Fc??R signaling as well. In contrast, Y551 sequestration was less a factor in determining potency against BCR signaling. We also found that Btk activity is regulated differentially in basophils and B cells. These results elucidate important determinants for Btk inhibitor potency against different signaling pathways and provide insight for designing new compounds with a broader inhibitory profile that will likely result in greater efficacy.
- External Link
- RCSB PDB
Ligand information
- HET
- 7G8
- Chain ID
- A
- HET Number
- 701
- Molecular Formula
- C26H22FN7O3
- Structure
-
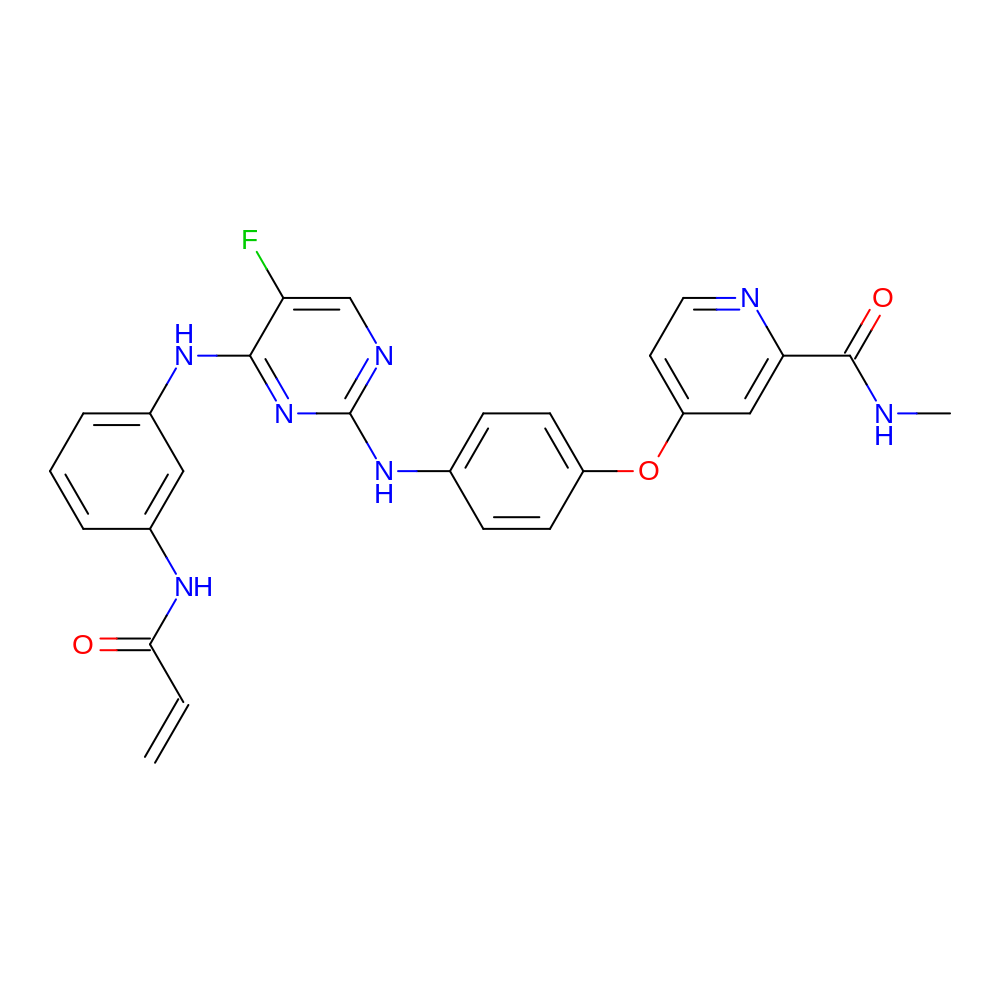
- IUPAC Name
- 4-[4-[[5-fluoro-4-[3-(prop-2-enoylamino)anilino]pyrimidin-2-yl]amino]phenoxy]-N-methyl-pyridine-2-carboxamide
- InChI
- InChI=1S/C26H22FN7O3/c1-3-23(35)31-17-5-4-6-18(13-17)32-24-21(27)15-30-26(34-24)33-16-7-9-19(10-8-16)37-20-11-12-29-22(14-20)25(36)28-2/h3-15H,1H2,2H3,(H,28,36)(H,31,35)(H2,30,32,33,34)
- InChI Key
- VVLHQJDAUIPZFH-UHFFFAOYSA-N
- Canonical SMILES
- CNC(=O)c1cc(ccn1)Oc2ccc(cc2)Nc3ncc(F)c(n3)Nc4cccc(c4)NC(=O)C=C
- Bioactivity data
- CI003308
Covalent Binding
- Warhead
- Michael Acceptor
- Reaction Mechanism
- Michael Addition
- Residue
- CYS : 481
- Residue Chain
- A
- Interactions
- Pharmacophore Model