4LL0
Target information
- RCSB PDB
- 4LL0
- Title
- EGFR L858R/T790M in complex with PD168393
- Method
- X-RAY DIFFRACTION
- Resolution
- 4
- Classification
- transferase/transferase inhibitor
- Organism
- Homo sapiens
- Protein
- Epidermal growth factor receptor (P00533) Looking for covalent inhibitors of this target ?
- Year
- 2013
- Publication Title
- Mechanism for activation of mutated epidermal growth factor receptors in lung cancer.
- Abstract
-
The initiation of epidermal growth factor receptor (EGFR) kinase activity proceeds via an asymmetric dimerization mechanism in which a 'donor' tyrosine kinase domain (TKD) contacts an 'acceptor' TKD, leading to its activation. In the context of a ligand-induced dimer, identical wild-type EGFR TKDs are thought to assume the donor or acceptor roles in a random manner. Here, we present biochemical reconstitution data demonstrating that activated EGFR mutants found in lung cancer preferentially assume the acceptor role when coexpressed with WT EGFR. Mutated EGFRs show enhanced association with WT EGFR, leading to hyperphosphorylation of the WT counterpart. Mutated EGFRs also hyperphosphorylate the related erythroblastic leukemia viral oncogene (ErbB) family member, ErbB-2, in a similar manner. This directional 'superacceptor activity' is particularly pronounced in the drug-resistant L834R/T766M mutant. A 4-? crystal structure of this mutant in the active conformation reveals an asymmetric dimer interface that is essentially the same as that in WT EGFR. Asymmetric dimer formation induces an allosteric conformational change in the acceptor subunit. Thus, superacceptor activity likely arises simply from a lower energetic cost associated with this conformational change in the mutant EGFR compared with WT, rather than from any structural alteration that impairs the donor role of the mutant. Collectively, these findings define a previously unrecognized mode of mutant-specific intermolecular regulation for ErbB receptors, knowledge of which could potentially be exploited for therapeutic benefit.
- External Link
- RCSB PDB
Ligand information
- HET
- YUN
- Chain ID
- B
- HET Number
- 1101
- Molecular Formula
- C17H13BrN4O
- Structure
-
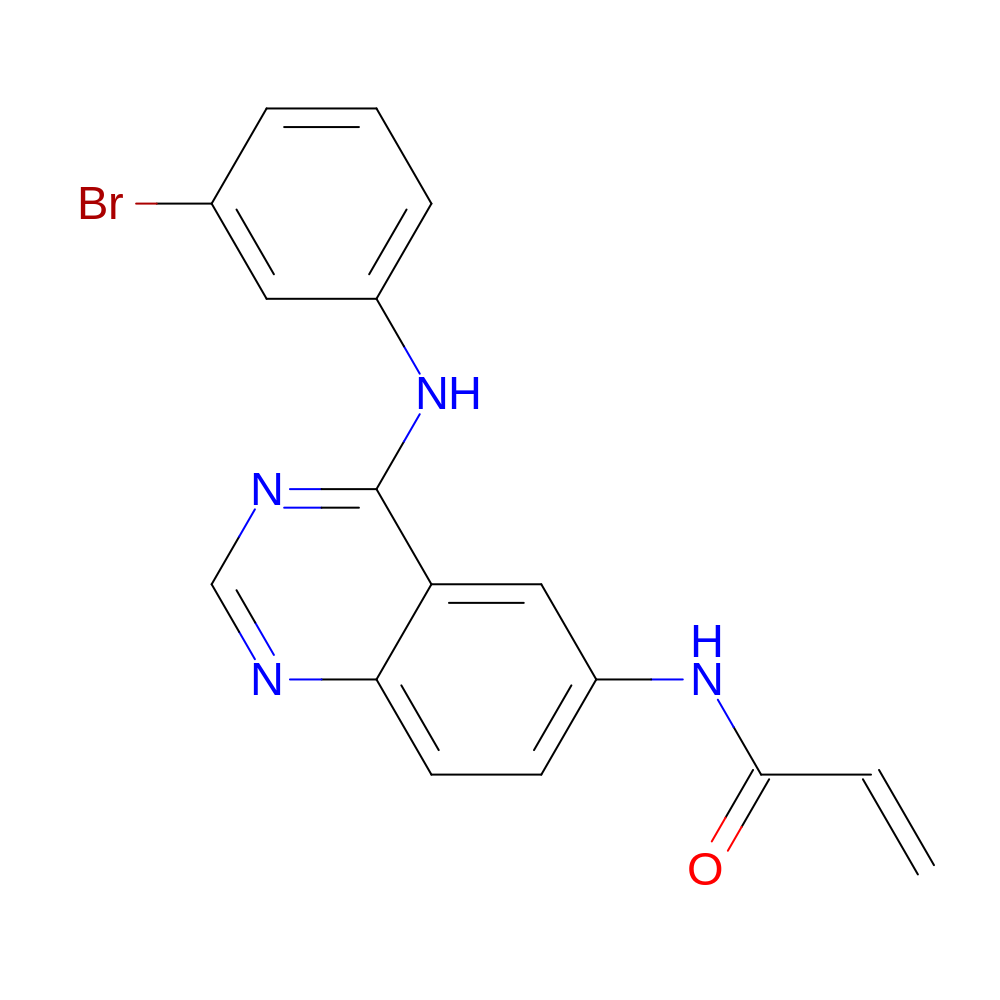
- IUPAC Name
- N-[4-(3-bromoanilino)quinazolin-6-yl]prop-2-enamide
- InChI
- InChI=1S/C17H13BrN4O/c1-2-16(23)21-13-6-7-15-14(9-13)17(20-10-19-15)22-12-5-3-4-11(18)8-12/h2-10H,1H2,(H,21,23)(H,19,20,22)
- InChI Key
- HTUBKQUPEREOGA-UHFFFAOYSA-N
- Canonical SMILES
- C=CC(=O)Nc1ccc2ncnc(Nc3cccc(Br)c3)c2c1
- Bioactivity data
- CI002137
Covalent Binding
- Warhead
- Michael Acceptor
- Reaction Mechanism
- Michael Addition
- Residue
- CYS : 797
- Residue Chain
- B
- Interactions
- Pharmacophore Model