4B55
Target information
- RCSB PDB
- 4B55
- Title
- Crystal Structure of the Covalent Adduct Formed between Mycobacterium marinum Aryalamine N-acetyltransferase and Phenyl vinyl ketone a derivative of Piperidinols
- Method
- X-RAY DIFFRACTION
- Resolution
- 2.7
- Classification
- TRANSFERASE
- Organism
- MYCOBACTERIUM MARINUM
- Protein
- Arylamine N-acetyltransferase Nat (B2HIZ6)
- Year
- 2012
- Publication Title
- Piperidinols that Show Anti-Tubercular Activity as Inhibitors of Arylamine N-Acetyltransferase: An Essential Enzyme for Mycobacterial Survival Inside Macrophages.
- Abstract
-
Latent M. tuberculosis infection presents one of the major obstacles in the global eradication of tuberculosis (TB). Cholesterol plays a critical role in the persistence of M. tuberculosis within the macrophage during latent infection. Catabolism of cholesterol contributes to the pool of propionyl-CoA, a precursor that is incorporated into cell-wall lipids. Arylamine N-acetyltransferase (NAT) is encoded within a gene cluster that is involved in the cholesterol sterol-ring degradation and is essential for intracellular survival. The ability of the NAT from M. tuberculosis (TBNAT) to utilise propionyl-CoA links it to the cholesterol-catabolism pathway. Deleting the nat gene or inhibiting the NAT enzyme prevents intracellular survival and results in depletion of cell-wall lipids. TBNAT has been investigated as a potential target for TB therapies. From a previous high-throughput screen, 3-benzoyl-4-phenyl-1-methylpiperidinol was identified as a selective inhibitor of prokaryotic NAT that exhibited antimycobacterial activity. The compound resulted in time-dependent irreversible inhibition of the NAT activity when tested against NAT from M. marinum (MMNAT). To further evaluate the antimycobacterial activity and the NAT inhibition of this compound, four piperidinol analogues were tested. All five compounds exert potent antimycobacterial activity against M. tuberculosis with MIC values of 2.3-16.9 µM. Treatment of the MMNAT enzyme with this set of inhibitors resulted in an irreversible time-dependent inhibition of NAT activity. Here we investigate the mechanism of NAT inhibition by studying protein-ligand interactions using mass spectrometry in combination with enzyme analysis and structure determination. We propose a covalent mechanism of NAT inhibition that involves the formation of a reactive intermediate and selective cysteine residue modification. These piperidinols present a unique class of antimycobacterial compounds that have a novel mode of action different from known anti-tubercular drugs.
- External Link
- RCSB PDB
Ligand information
- HET
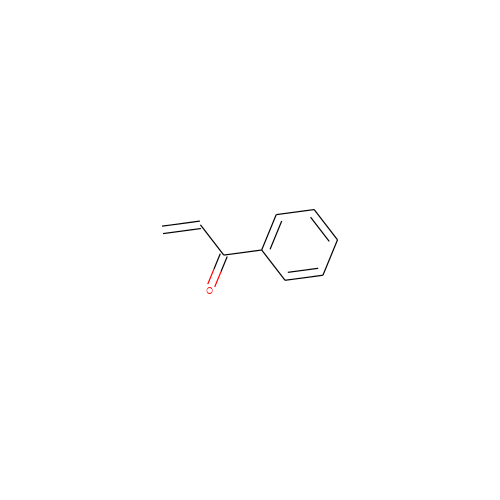
- P18
- Chain ID
- A
- HET Number
- 1276
- Molecular Formula
- C9H8O
- Structure
-
- IUPAC Name
- 1-phenylprop-2-en-1-one
- InChI
- InChI=1S/C9H8O/c1-2-9(10)8-6-4-3-5-7-8/h2-7H,1H2
- InChI Key
- KUIZKZHDMPERHR-UHFFFAOYSA-N
- Canonical SMILES
- C=CC(=O)c1ccccc1
- Bioactivity data
- CI000036
Covalent Binding
- Warhead
- Michael Acceptor
- Reaction Mechanism
- Michael Addition
- Residue
- CYS : 70
- Residue Chain
- A
- Interactions
- Pharmacophore Model