3LOK
Target information
- RCSB PDB
- 3LOK
- Title
- Drug resistant cSrc kinase domain in complex with covalent inhibitor PD168393
- Method
- X-RAY DIFFRACTION
- Resolution
- 2.48
- Classification
- TRANSFERASE/TRANSFERASE INHIBITOR
- Organism
- Gallus gallus
- Protein
- Proto-oncogene tyrosine-protein kinase Src (P00523) Looking for covalent inhibitors of this target ?
- Year
- 2010
- Publication Title
- Characterization of irreversible kinase inhibitors by directly detecting covalent bond formation: a tool for dissecting kinase drug resistance
- Abstract
-
Targeting protein kinases in cancer therapy with irreversible small-molecule inhibitors is moving to the forefront of kinase-inhibitor research and is thought to be an effective means of overcoming mutation-associated drug resistance in epidermal growth factor receptor kinase (EGFR). We generated a detection technique that allows direct measurements of covalent bond formation without relying on kinase activity, thereby allowing the straightforward investigation of the influence of steric clashes on covalent inhibitors in different resistant kinase mutants. The obtained results are discussed together with structural biology and biochemical studies of catalytic activity in both wild-type and gatekeeper mutated kinase variants to draw conclusions about the impact of steric hindrance and increased catalytic activity in drug-resistant kinase variants.
- External Link
- RCSB PDB
Ligand information
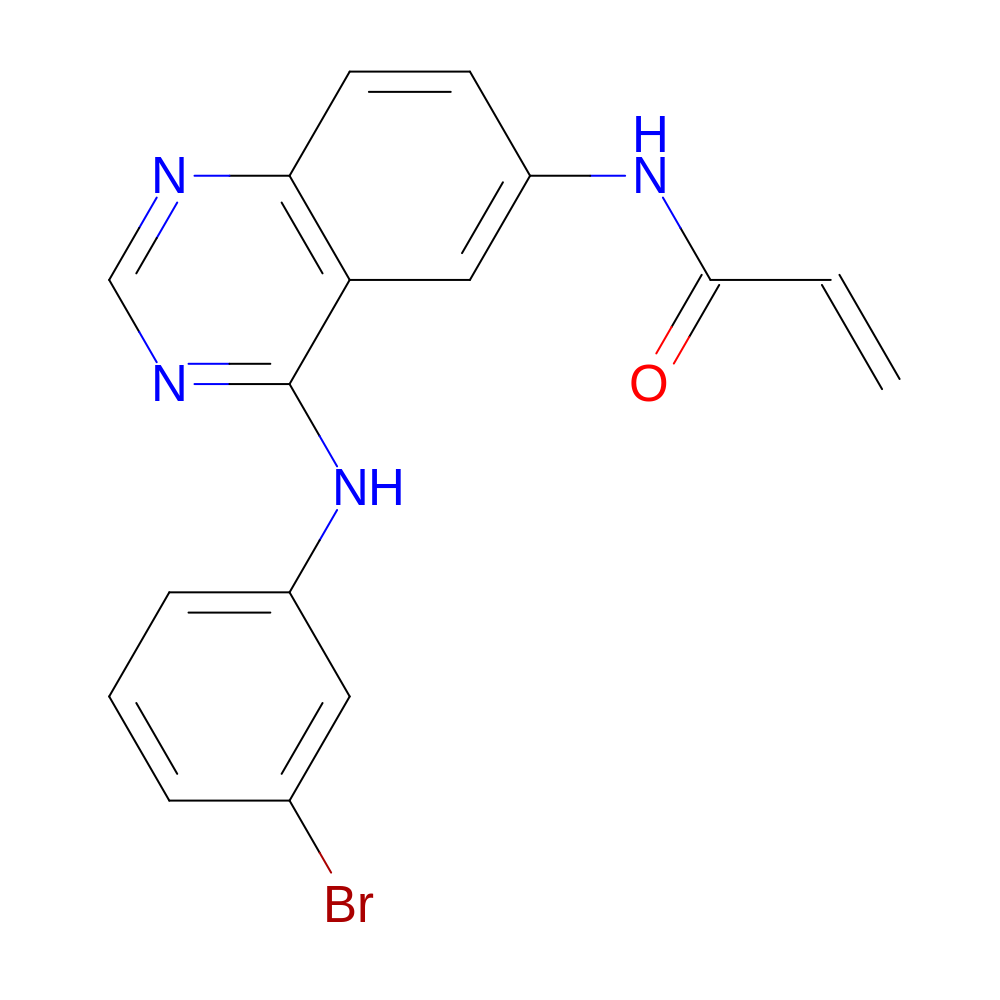
- HET
- DJK
- Chain ID
- A
- HET Number
- 1345
- Molecular Formula
- C17H13BrN4O
- Structure
-
- IUPAC Name
- N-[4-(3-bromoanilino)quinazolin-6-yl]prop-2-enamide
- InChI
- InChI=1S/C17H13BrN4O/c1-2-16(23)21-13-6-7-15-14(9-13)17(20-10-19-15)22-12-5-3-4-11(18)8-12/h2-10H,1H2,(H,21,23)(H,19,20,22)
- InChI Key
- HTUBKQUPEREOGA-UHFFFAOYSA-N
- Canonical SMILES
- C=CC(=O)Nc(c1)ccc(c12)ncnc2Nc3cccc(Br)c3
- Bioactivity data
- CI002137
Covalent Binding
- Warhead
- Michael Acceptor
- Reaction Mechanism
- Michael Addition
- Residue
- CYS : 345
- Residue Chain
- A
- Interactions
- Pharmacophore Model