3IKA
Target information
- RCSB PDB
- 3IKA
- Title
- Crystal Structure of EGFR 696-1022 T790M Mutant Covalently Binding to WZ4002
- Method
- X-RAY DIFFRACTION
- Resolution
- 2.9
- Classification
- TRANSFERASE
- Organism
- Homo sapiens
- Protein
- Epidermal growth factor receptor (P00533) Looking for covalent inhibitors of this target ?
- Year
- 2009
- Publication Title
- Novel mutant-selective EGFR kinase inhibitors against EGFR T790M.
- Abstract
-
The clinical efficacy of epidermal growth factor receptor (EGFR) kinase inhibitors in EGFR-mutant non-small-cell lung cancer (NSCLC) is limited by the development of drug-resistance mutations, including the gatekeeper T790M mutation. Strategies targeting EGFR T790M with irreversible inhibitors have had limited success and are associated with toxicity due to concurrent inhibition of wild-type EGFR. All current EGFR inhibitors possess a structurally related quinazoline-based core scaffold and were identified as ATP-competitive inhibitors of wild-type EGFR. Here we identify a covalent pyrimidine EGFR inhibitor by screening an irreversible kinase inhibitor library specifically against EGFR T790M. These agents are 30- to 100-fold more potent against EGFR T790M, and up to 100-fold less potent against wild-type EGFR, than quinazoline-based EGFR inhibitors in vitro. They are also effective in murine models of lung cancer driven by EGFR T790M. Co-crystallization studies reveal a structural basis for the increased potency and mutant selectivity of these agents. These mutant-selective irreversible EGFR kinase inhibitors may be clinically more effective and better tolerated than quinazoline-based inhibitors. Our findings demonstrate that functional pharmacological screens against clinically important mutant kinases represent a powerful strategy to identify new classes of mutant-selective kinase inhibitors.
- External Link
- RCSB PDB
Ligand information
- HET
- 0UN
- Chain ID
- A
- HET Number
- 1797
- Molecular Formula
- C25H27ClN6O3
- Structure
-
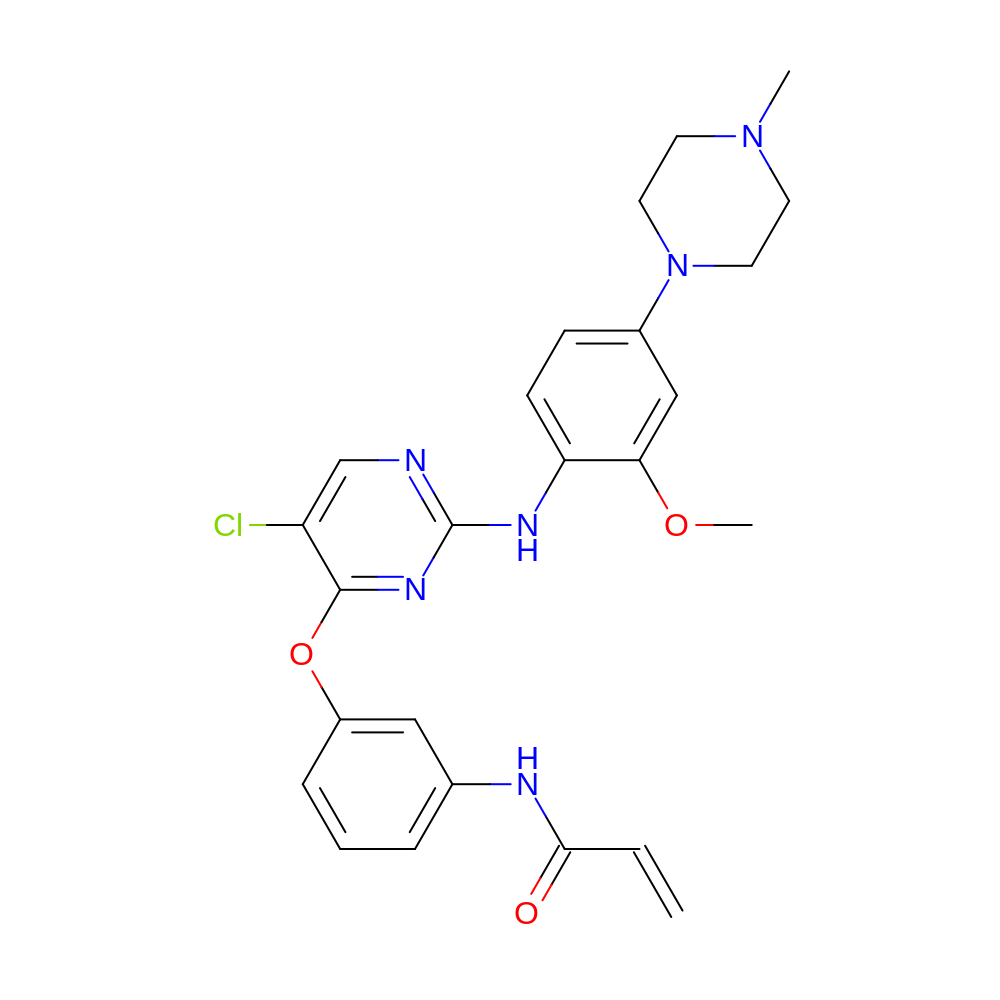
- IUPAC Name
- N-[3-[5-chloro-2-[2-methoxy-4-(4-methylpiperazin-1-yl)anilino]pyrimidin-4-yl]oxyphenyl]prop-2-enamide
- InChI
- InChI=1S/C25H27ClN6O3/c1-4-23(33)28-17-6-5-7-19(14-17)35-24-20(26)16-27-25(30-24)29-21-9-8-18(15-22(21)34-3)32-12-10-31(2)11-13-32/h4-9,14-16H,1,10-13H2,2-3H3,(H,28,33)(H,27,29,30)
- InChI Key
- ITTRLTNMFYIYPA-UHFFFAOYSA-N
- Canonical SMILES
- C=CC(=O)Nc1cccc(Oc2nc(Nc3ccc(N4CCN(C)CC4)cc3OC)ncc2Cl)c1
- Bioactivity data
- CI000932
Covalent Binding
- Warhead
- Michael Acceptor
- Reaction Mechanism
- Michael Addition
- Residue
- CYS : 797
- Residue Chain
- A
- Interactions
- Pharmacophore Model