3I03
Target information
- RCSB PDB
- 3I03
- Title
- Crystal structure of bothropstoxin-I chemically modified by p-bromophenacyl bromide (BPB) - monomeric form at a high resolution
- Method
- X-RAY DIFFRACTION
- Resolution
- 1.48
- Classification
- TOXIN
- Organism
- Bothrops jararacussu
- Protein
- Basic phospholipase A2 homolog bothropstoxin-I (Q90249)
- Year
- 2009
- Publication Title
- Comparison between apo and complexed structures of bothropstoxin-I reveals the role of Lys122 and Ca(2+)-binding loop region for the catalytically inactive Lys49-PLA(2)s.
- Abstract
-
Phospholipases A(2) (Asp49-PLA(2)s) are enzymes responsible for cellular membrane disruption through Ca(2+)-dependent hydrolysis of phospholipids. A class of these proteins (Lys49-PLA(2)s) does not show catalytic activity but can exert a pronounced local myotoxic effect that is not neutralized by serum therapy. In this work, we present five structures of Lys49-PLA(2)s from snakes of the Bothrops genus in apo form, complexed with PEG molecules and chemically modified by p-bromofenacil bromide (BPB), a classic inhibitor of PLA(2). We present herein an extensive structural analysis including: (i) the function of hydrophobic long-chain molecules as Lys49-PLA(2)s inhibitors, (ii) the role of Lys122, previously indicated as being responsible for Lys49-PLA(2)s catalytic inactivity and, (iii) a structural comparison of the Ca(2+)-binding loop region between Lys49 and Asp49-PLA(2)s. The Lys122 analysis of 30 different monomers for apo and complexed Lys49-PLA(2)s structures shows that this residue is very flexible and may bind to different carboxyl groups giving stability to the crystal structures. The structural comparisons of the Ca(2+)-binding loop region between Lys49 and Asp49-PLA(2)s reveal the importance of the Tyr28 residue conservation in Asp49-PLA(2)s to the integrity of this loop. The Tyr28 residue stabilizes this region by an interaction with Gly35 residue. In Lys49-PLA(2)s and low-catalytic Asp49-PLA(2)s this interaction does not occur, preventing the binding of Ca(2+).
- External Link
- RCSB PDB
Ligand information
- HET
- PBP
- Chain ID
- A
- HET Number
- 201
- Molecular Formula
- C8H6Br2O
- Structure
-
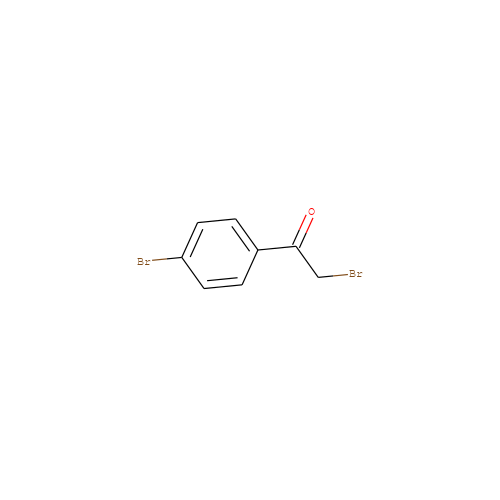
- IUPAC Name
- 2-bromo-1-(4-bromophenyl)ethanone
- InChI
- InChI=1S/C8H6Br2O/c9-5-8(11)6-1-3-7(10)4-2-6/h1-4H,5H2
- InChI Key
- FKJSFKCZZIXQIP-UHFFFAOYSA-N
- Canonical SMILES
- Brc1ccc(cc1)C(=O)CBr
- Bioactivity data
- CI002419
Covalent Binding
- Warhead
- Halohydrocarbon
- Reaction Mechanism
- Nucleophilic Substitution
- Residue
- HIS : 47
- Residue Chain
- A
- Interactions
- Pharmacophore Model