2QQ7
Target information
- RCSB PDB
- 2QQ7
- Title
- Crystal structure of drug resistant SRC kinase domain with irreversible inhibitor
- Method
- X-RAY DIFFRACTION
- Resolution
- 2.38
- Classification
- TRANSFERASE
- Organism
- Gallus gallus
- Protein
- Proto-oncogene tyrosine-protein kinase Src (P00523) Looking for covalent inhibitors of this target ?
- Year
- 2007
- Publication Title
- Structural insights into how irreversible inhibitors can overcome drug resistance in EGFR.
- Abstract
-
Resistance to kinase-targeted cancer drugs has recently been linked to a single point mutation in the ATP binding site of the kinase. In EGFR, the crucial Thr790 gatekeeper residue is mutated to a Met and prevents reversible ATP competitive inhibitors from binding. Irreversible 4-(phenylamino)quinazolines have been shown to overcome this drug resistance and are currently in clinical trials. In order to obtain a detailed structural understanding of how irreversible inhibitors overcome drug resistance, we used Src kinase as a model system for drug resistant EGFR-T790M. We report the first crystal structure of a drug resistant kinase in complex with an irreversible inhibitor. This 4-(phenylamino)quinazoline inhibits wild type and drug resistant EGFR in vitro at low nM concentrations. The co-crystal structure of drug resistant cSrc-T338M kinase domain provides the structural basis of this activity.
- External Link
- RCSB PDB
Ligand information
- HET
- SR2
- Chain ID
- A
- HET Number
- 1345
- Molecular Formula
- C20H20BrN5O
- Structure
-
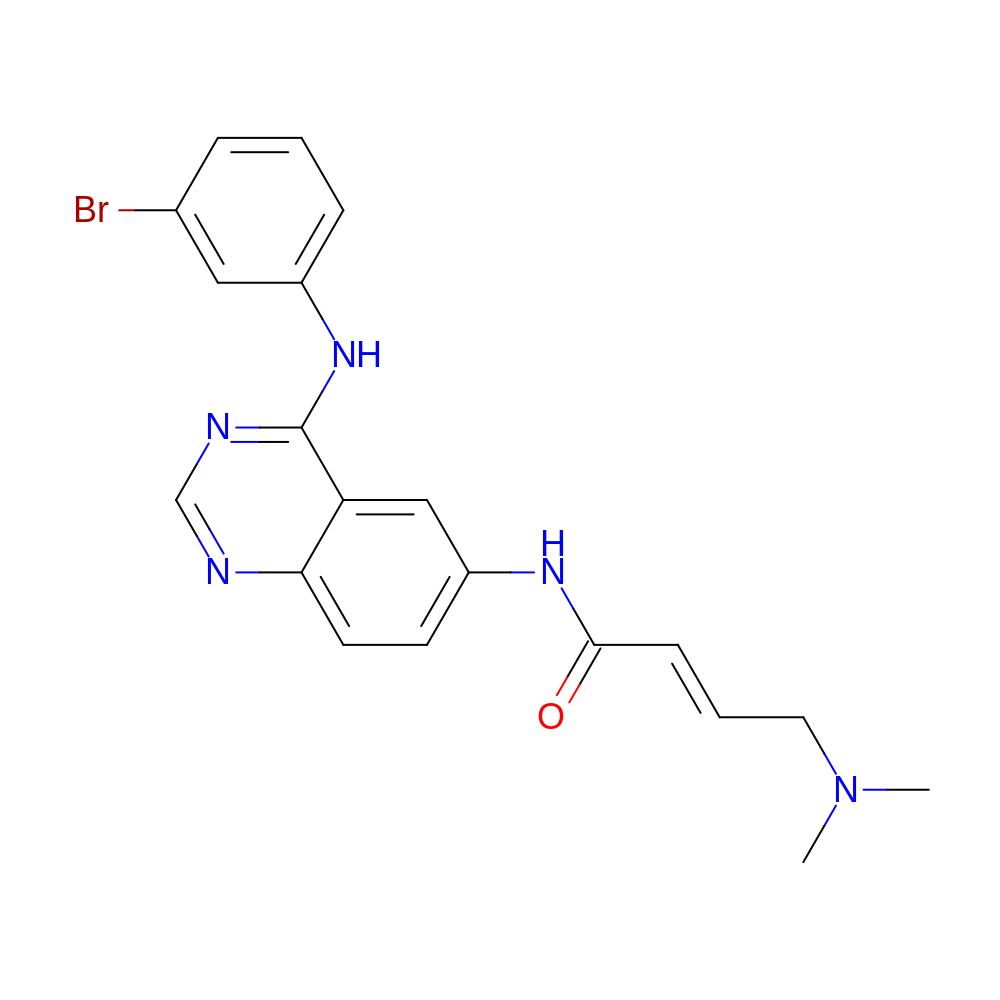
- IUPAC Name
- (E)-N-[4-(3-bromoanilino)quinazolin-6-yl]-4-(dimethylamino)but-2-enamide
- InChI
- InChI=1S/C20H20BrN5O/c1-26(2)10-4-7-19(27)24-16-8-9-18-17(12-16)20(23-13-22-18)25-15-6-3-5-14(21)11-15/h3-9,11-13H,10H2,1-2H3,(H,24,27)(H,22,23,25)/b7-4+
- InChI Key
- ZCIXBBSRVLSRJQ-QPJJXVBHSA-N
- Canonical SMILES
- CN(C)C\C=C\C(=O)Nc1ccc2ncnc(Nc3cccc(Br)c3)c2c1
- Bioactivity data
- CI002234
Covalent Binding
- Warhead
- Michael Acceptor
- Reaction Mechanism
- Michael Addition
- Residue
- CYS : 345
- Residue Chain
- A
- Interactions
- Pharmacophore Model